I try to use cutlass byoc, and the finally execution seems some input lost.
import imp
import tvm
from tvm import relay
import numpy as np
from tvm.relay.op.contrib import partition_for_cutlass
from tvm.contrib.cutlass.build import tune_cutlass_kernels, build_cutlass_kernels
def get_data(M, N, K, out_dtype="float16"):
np_data = np.random.uniform(-1, 1, (M, K)).astype("float16")
np_weight = np.random.uniform(-1, 1, (N, K)).astype("float16")
np_bias = np.random.uniform(-1, 1, (N,)).astype(out_dtype)
return np_data, np_weight, np_bias
def get_output(rt_mod, names, inputs):
# import pdb
# pdb.set_trace()
# for name, inp in zip(names, inputs):
# rt_mod.set_input(name, inp)
rt_mod.run()
return rt_mod.get_output(0).asnumpy()
def profile_and_build(mod, params, sm, tmp_dir="./tmp", lib_path="compile.so"):
mod = partition_for_cutlass(mod)
mod, num_cutlass_partition = tune_cutlass_kernels(mod, sm, use_multiprocessing=False, tmp_dir=tmp_dir)
with tvm.transform.PassContext(opt_level=3):
lib = relay.build(mod, target="cuda", params=params)
print("finaly output ir_mod", lib.ir_mod)
lib = build_cutlass_kernels(lib, sm, tmp_dir, lib_path)
dev = tvm.device("cuda", 0)
rt_mod = tvm.contrib.graph_executor.GraphModule(lib["default"](dev))
return rt_mod, dev, num_cutlass_partition
def get_dense(M, N, K, output_type="float16"):
data = relay.var("data", shape=(M, K), dtype="float16")
weight = relay.var("weight", shape=(N, K), dtype="float16")
scale = relay.var("scale", shape=(M, K), dtype="float16")
bias = relay.var("bias", shape=(N, ), dtype=output_type)
data = relay.add(data, scale)
dense_output = relay.nn.dense(data, weight, out_dtype=output_type)
bias_add_output = relay.nn.bias_add(dense_output, bias)
relu_output = relay.nn.relu(bias_add_output)
return relu_output
def test_dense():
M = 16384
N =768
K = 3072
sm = 75
dense_output = get_dense(M, N, K, "float32")
mod = tvm.IRModule.from_expr(dense_output)
typ = relay.transform.InferType()(mod)["main"].body.checked_type
out_dtype = typ.dtype
np_data = np.random.uniform(-1, 1, (M, K)).astype("float16")
np_weight = np.random.uniform(-1, 1, (N, K)).astype("float16")
np_scale = np.random.uniform(-1, 1, (M, K)).astype("float16")
np_bias = np.random.uniform(-1, 1, (N,)).astype(out_dtype)
params = {"data": np_data, "weight": np_weight, "bias": np_bias, "scale": np_scale}
rt_mod, dev, num_partition = profile_and_build(mod, params, sm)
data = tvm.nd.array(np_data, device=dev)
weight = tvm.nd.array(np_weight, device=dev)
bias = tvm.nd.array(np_bias, device=dev)
scale = tvm.nd.array(np_scale, device=dev)
out = get_output(rt_mod, ["data", "weight", "bias", "scale"], [data, weight, bias, scale])
if __name__ == "__main__":
test_dense()
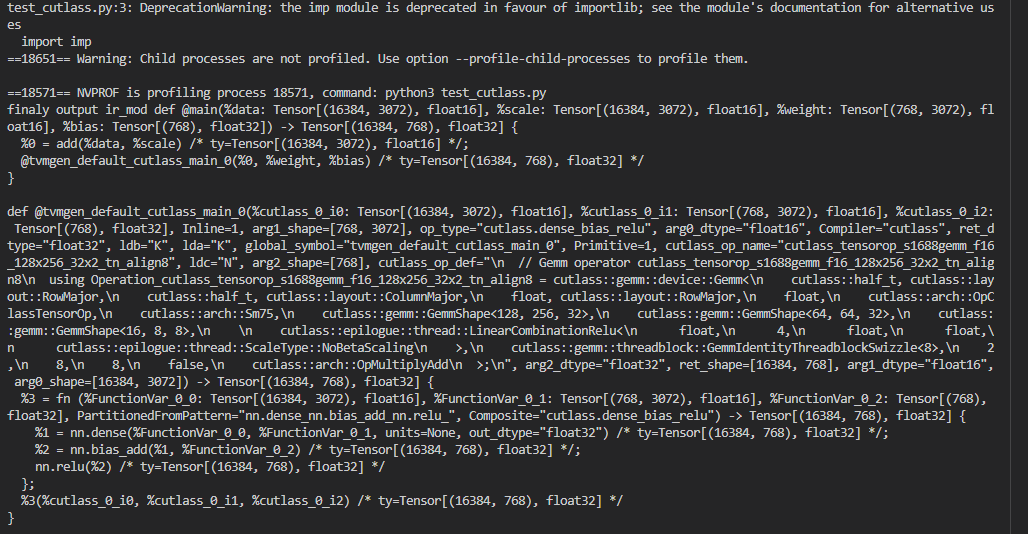
the output ir mod may like:
so we can the in ir mod the add op does exist, but if i run the lib.
only execute the gemm. The add op seems lost.
and the input is not data, weight ,bias, and scale, but p0, p1, p2.